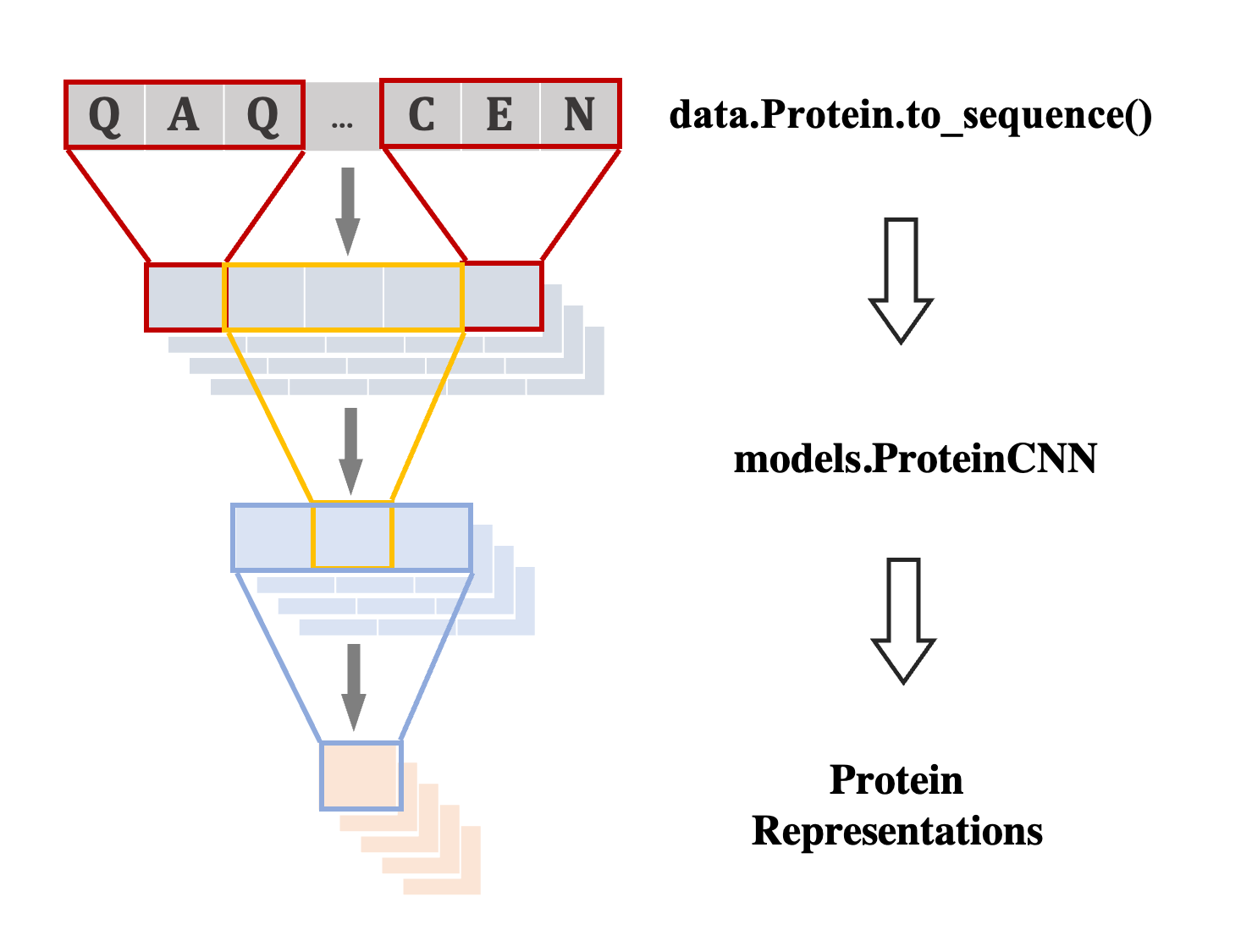
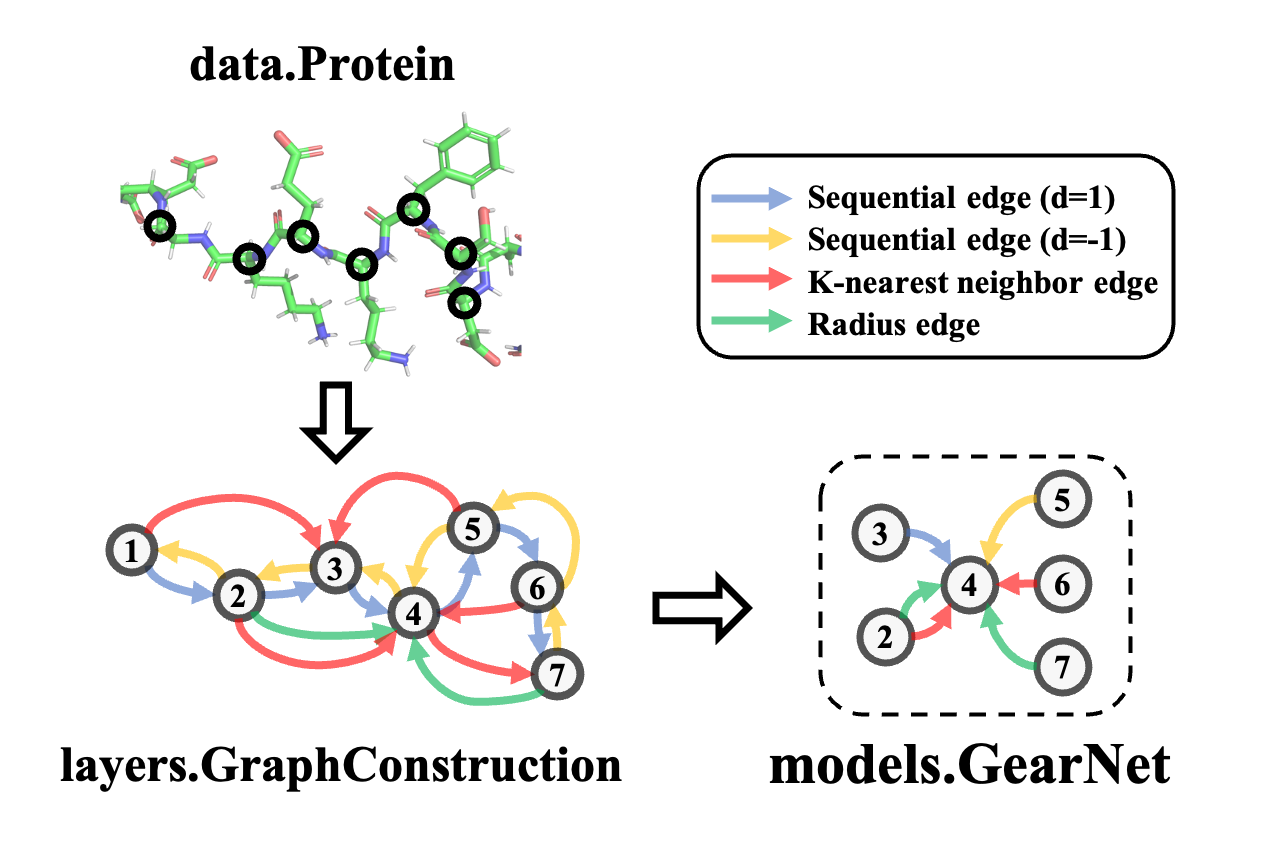
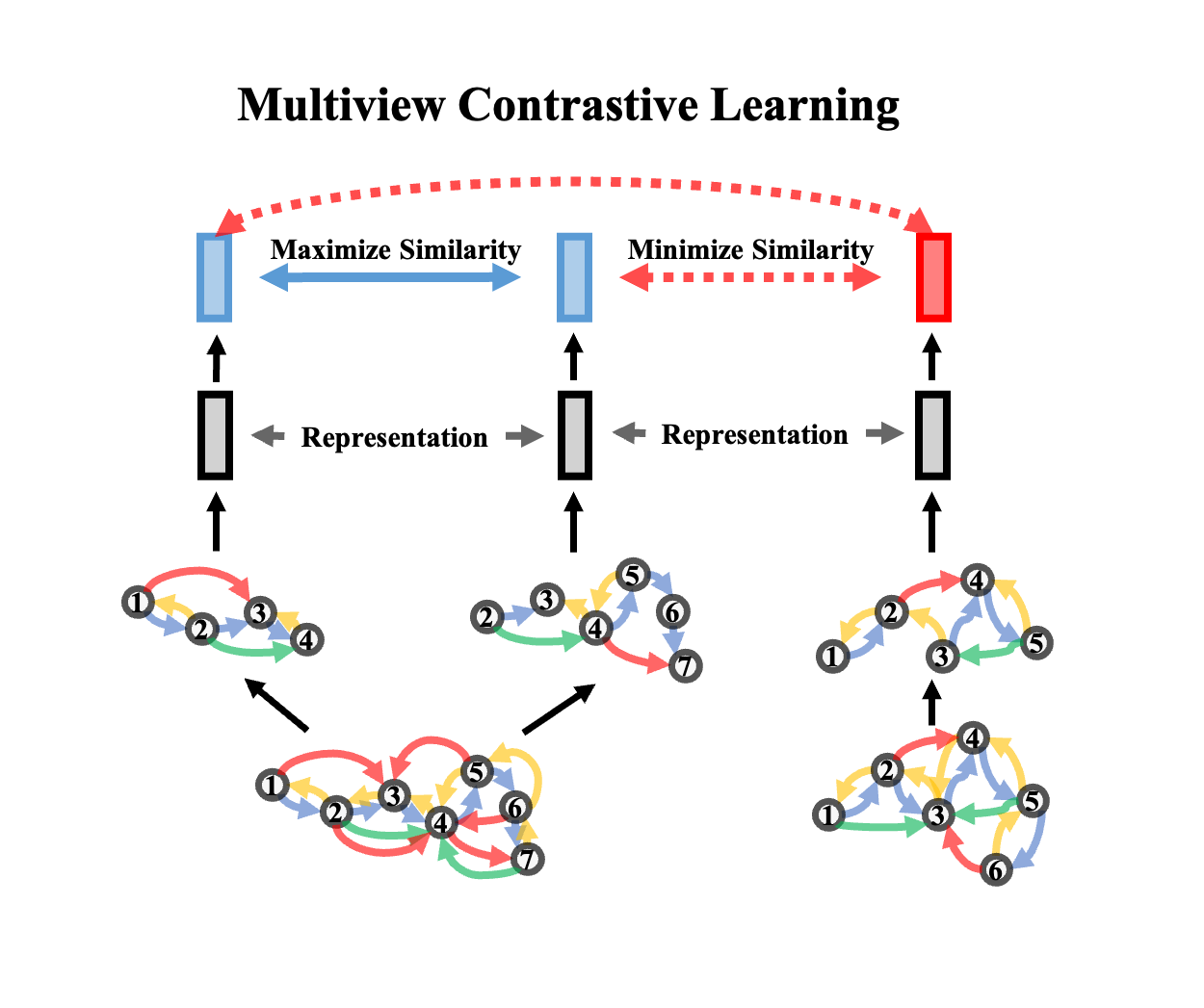
Protein machine learning accessible to everyone
TorchProtein is a machine learning library for protein science, built on top of TorchDrug. It provides representation learning models for both protein sequences and structures, as well as fundamental protein tasks like function prediction, structure prediction. Taking the advantage of TorchDrug, it is also easy to reuse abundant models from small molecules for proteins, or solve protein-molecule tasks such as binding affinity prediction.
Available as a part of TorchDrug.
$ pip install torchdrug
Why TorchProtein?
TorchProtein is an open source library for protein representation learning. It encapsulates common protein machine learning demands in human-friendly data structures, models and tasks, to ease the process of building applications on proteins.
Unified Data Structures
Provide intuitive interface for mainuplating proteins and a large collection of common datasets for development.

Flexible Building Blocks
Boost model construction with generic building blocks and dynamic graph construction tailored to proteins.
Extensive Benchmarks
Comprehensively compare different representation learning models for protein structure and sequence understanding.

A Protein ML Model Zoo
Power the performance with an abundant model zoo of protein representation learning models.